Analzying Pupil Labs Neon Data With GazeR
🎃 Happy Halloween, goils and boils! 👻
I have a spooktacular blog post for you today—one that dives into the mysterious world of pupil data from Pupil Labs Neon glasses. We’re currently using the Neon in the classroom to study mind wandering and attention. However, these mobile eye trackers aren’t just for field studies—they shine in the lab too.
To demonstrate this, I created a simple PsychoPy experiment that interfaces seamlessly with the Neon (you can find it here: https://osf.io/txz59/overview ). In the task, participants view a bright sun for 30 seconds followed by a dark patch for another 30 seconds. The pupil responds to basic visual features like brightness—constricting in light and dilating in the dark.
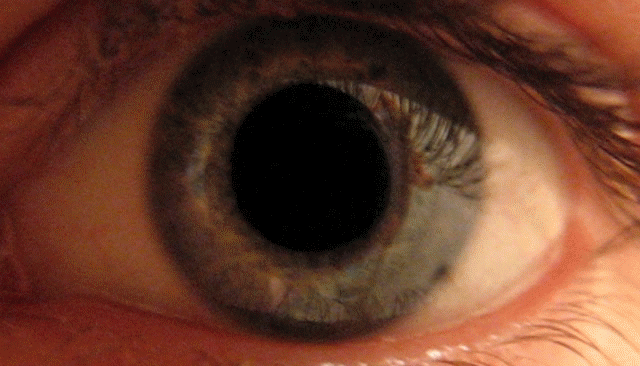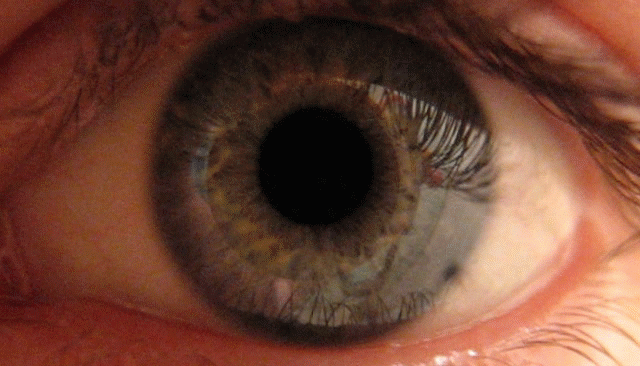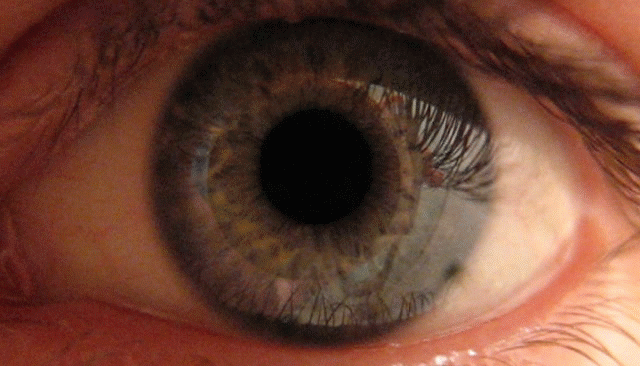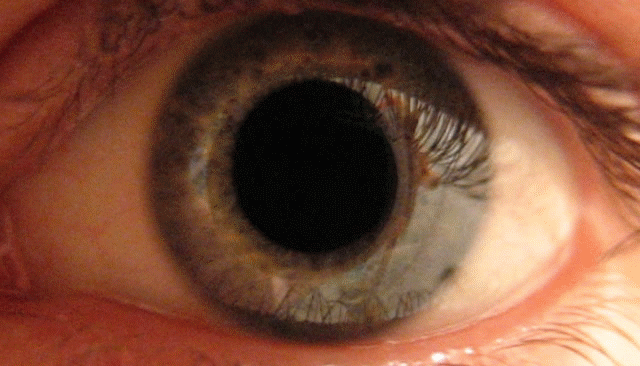
In this post, I’ll show how the Neon glasses can capture these pupillary dynamics and how you can use my R package {gazeR} to preprocess pupil data collected from Pupil Labs devices.
gazeR Pupil labs Functions
Once you collect data with Pupil Labs Neon, your recordings live in Pupil Cloud. Export the Time Series for each recording (CSV export). After the export finishes, you’ll have a folder per participant/recording containing multiple CSV files.
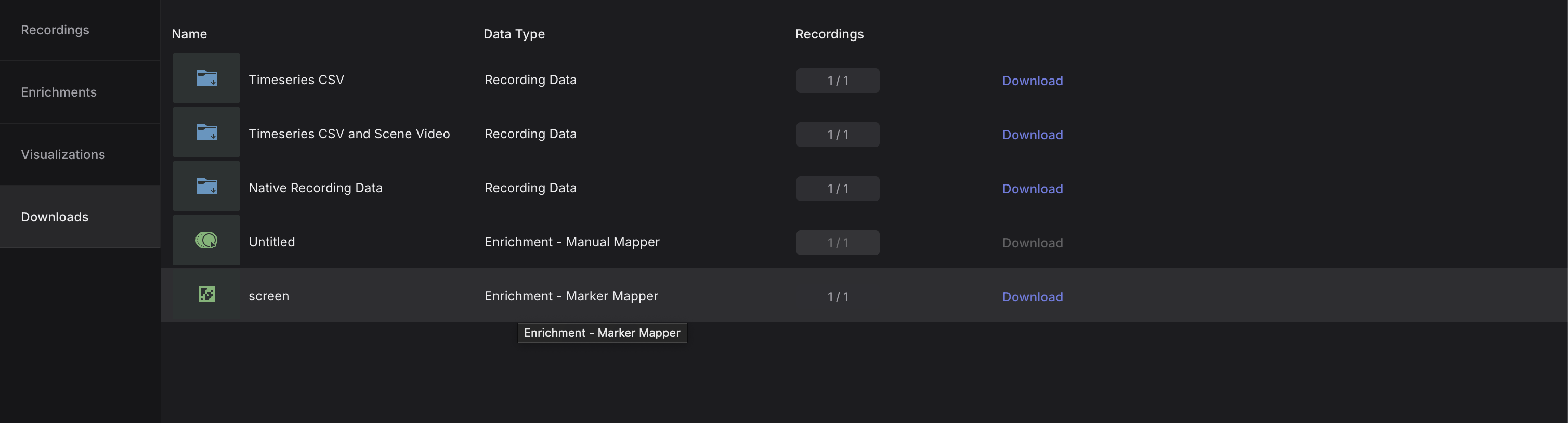
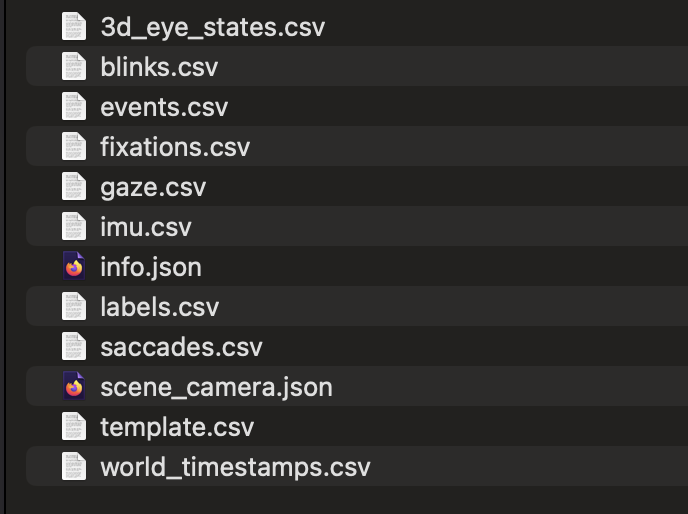
From each participant folder, we use exactly three files:
gaze.csv — gaze samples (timestamps, x/y pixels, fixations, blinks)
3d_eye_states.csv — pupil diameters (left/right, in mm)
events.csv — experiment events/messages (e.g., trial markers)
I created two new functions to read and process this data from Pupil Labs:
parse_pl() and process_all_subjects_PL(). Both work in tandem to prepare Neon data for use with {gazeR}.
What the functions do
parse_pl(subject_dir, aoi = FALSE, start_mode = c("any","exact"), start_messages = NULL, max_event_lag_ms = 20)
Processes one participant:
Reads the three CSVs and converts timestamps to milliseconds.
Joins pupil data to gaze samples.
Aligns each non-recording event (events.csv) to the nearest gaze row (within max_event_lag_ms).
If aoi=TRUE, it can read in the surface or AOI files. Here we used the marker mapper to map gaze on the screen of the laptop I used to record the data.
Creates trial indices:
start_mode = “any” → any non-empty message (excluding recording.begin/.end) starts a new trial.
start_mode = “exact” → only messages listed in start_messages start a new trial (robust to case/whitespace/hyphen differences).
Resets time to 0 at the first row of each trial.
Returns a tidy tibble ready for {gazeR}:
subject, trial, time, x, y, pupil, blink, message.
If you read in surface data, it will include whether gaze was found on the surface or not as well as x,y surface coordinates.
process_all_subjects_PL(root_dir, output_dir = file.path(root_dir, "processed"), ...)
Batch-processes all immediate subfolders of root_dir using parse_pl(), writes one CSV per subject plus a combined files
-
Per-subject files:
{output_dir}/{SUBJECT}_processed.csCombined file:
{output_dir}/all_subjects_processed.csv
Any additional arguments (...) are passed straight to parse_pl() (e.g., start_mode, start_messages, max_event_lag_ms).
Example Dataset
Let’s read in the dataset created from the above functions.
We will load in {gazeR} and needed libraries.
| subject | trial | time | x | y | pupil | blink | message |
|---|---|---|---|---|---|---|---|
| 2025-10-31_12-48-26-74964918 | 1 | 0.000000 | 813.938 | 606.636 | 4.24975 | FALSE | trial-started-light |
| 2025-10-31_12-48-26-74964918 | 1 | 5.005127 | 815.400 | 608.674 | 4.21485 | FALSE | |
| 2025-10-31_12-48-26-74964918 | 1 | 9.994873 | 815.163 | 607.715 | 4.23690 | FALSE | |
| 2025-10-31_12-48-26-74964918 | 1 | 14.994629 | 814.249 | 606.579 | 4.25715 | FALSE | |
| 2025-10-31_12-48-26-74964918 | 1 | 19.994629 | 812.541 | 607.620 | 4.22035 | FALSE | |
| 2025-10-31_12-48-26-74964918 | 1 | 24.994873 | 815.149 | 607.083 | 4.20380 | FALSE |
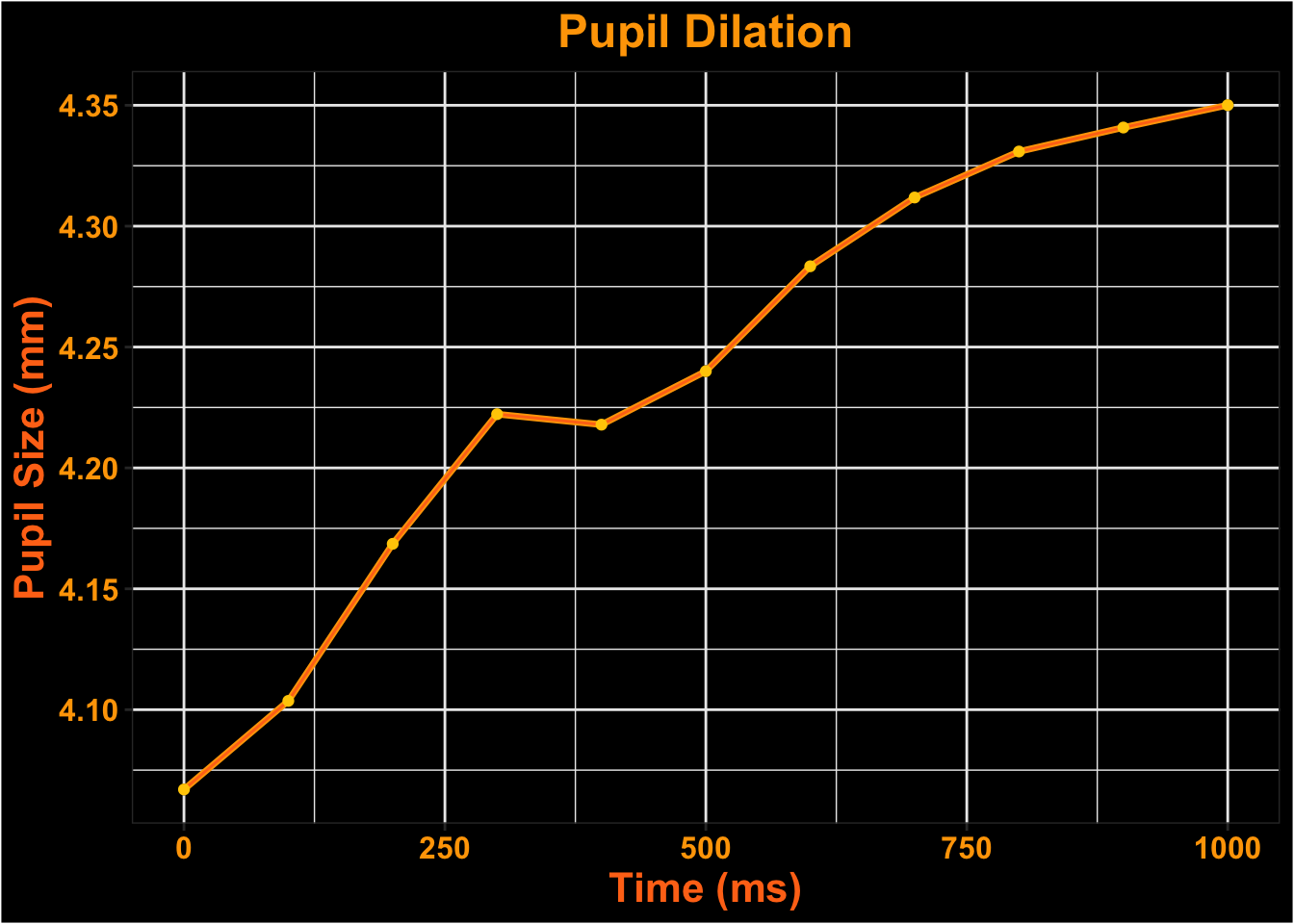
Let’s take a look at the data we have.
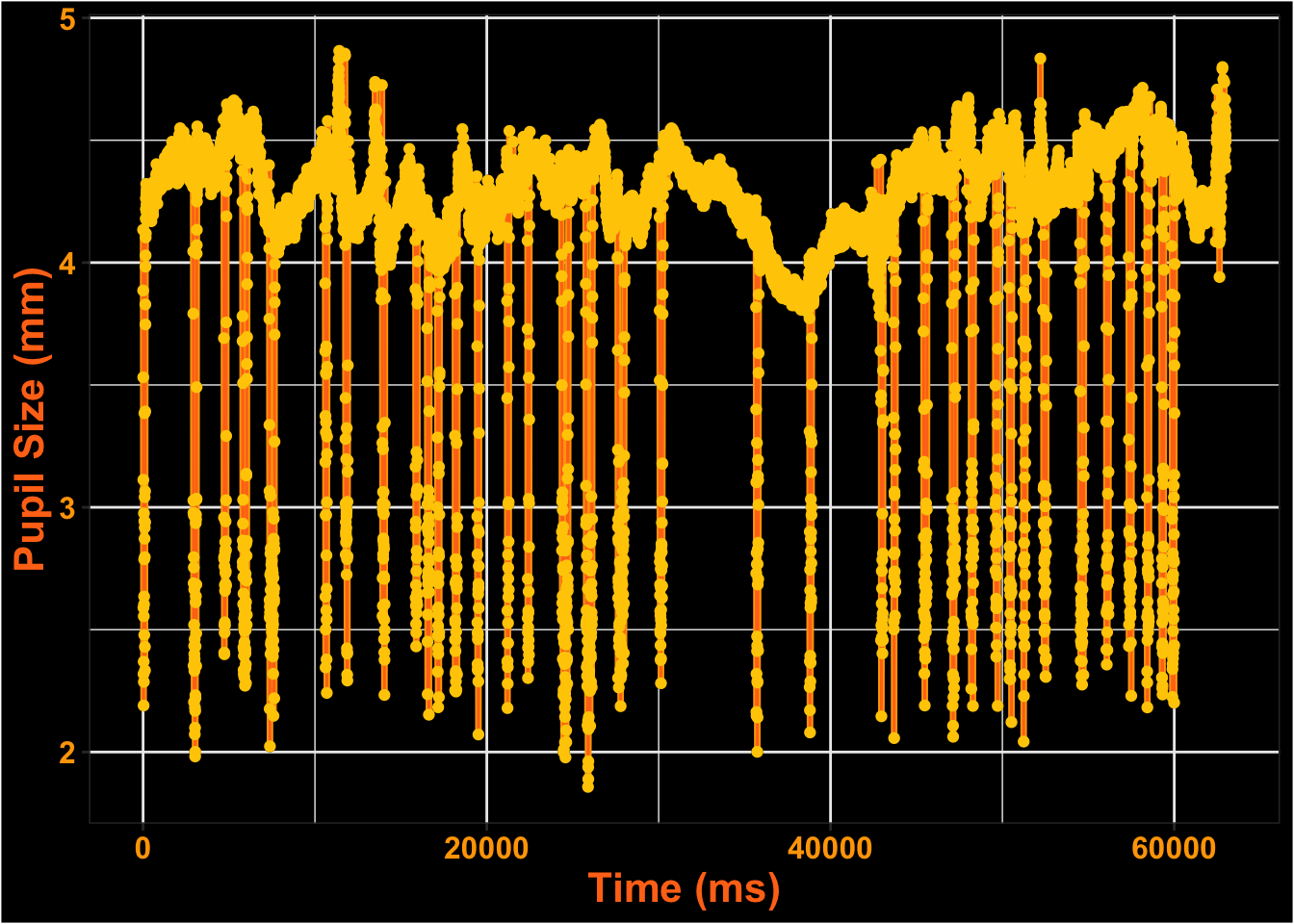
This is what the pupil data looks like for the entire time course.
Extending Blinks
We see dips in the pupil signal—these are most likely caused by blinks. Pupil Labs includes a blink detection algorithm, which we’ll use here. We first set pupil size values corresponding to blinks to NA, then use extend_blinks()to extend blinks 100 ms forward and backward in time.
Interpolate blinks
Next, we linearly interpolate over blinks and smooth the data using a 5-point moving average.
# Smooth and Interpolate
smooth_interp <- smooth_interpolate_pupil(pup_extend, pupil="pupil", extendpupil="extendpupil", extendblinks=TRUE, step.first="smooth", maxgap=Inf, type="linear", hz=200, n=5)Then, calculate missing data and remove trials with more than 50% missing data.
pup_missing<-count_missing_pupil(smooth_interp, missingthresh = .5)Unlikely Pupil Sizes
Now, keep only plausible pupil diameters between 2 mm and 9 mm.
pup_outliers<-pup_missing |>
dplyr::filter (pup_interp >= 2, pup_interp <= 9)MAD
Get rid of artifacts we might have missed during some earlier steps.
Onset
Let’s only look from the start of the trial until 1000 ms
Downsample
Downsample the time-course to 100 ms.
#downsample
timebins1<- downsample_gaze(pupil_onset, bin.length=100, pupil="pup_interp", timevar = "time", aggvars = c("subject", "trial", "timebins"), type="pupil")
kable(head(timebins1))| subject | trial | timebins | aggbaseline |
|---|---|---|---|
| 2025-10-31_12-48-26-74964918 | 1 | 0 | 4.233871 |
| 2025-10-31_12-48-26-74964918 | 1 | 100 | 4.251030 |
| 2025-10-31_12-48-26-74964918 | 1 | 200 | 4.264931 |
| 2025-10-31_12-48-26-74964918 | 1 | 300 | 4.296879 |
| 2025-10-31_12-48-26-74964918 | 1 | 400 | 4.323454 |
| 2025-10-31_12-48-26-74964918 | 1 | 500 | 4.247790 |
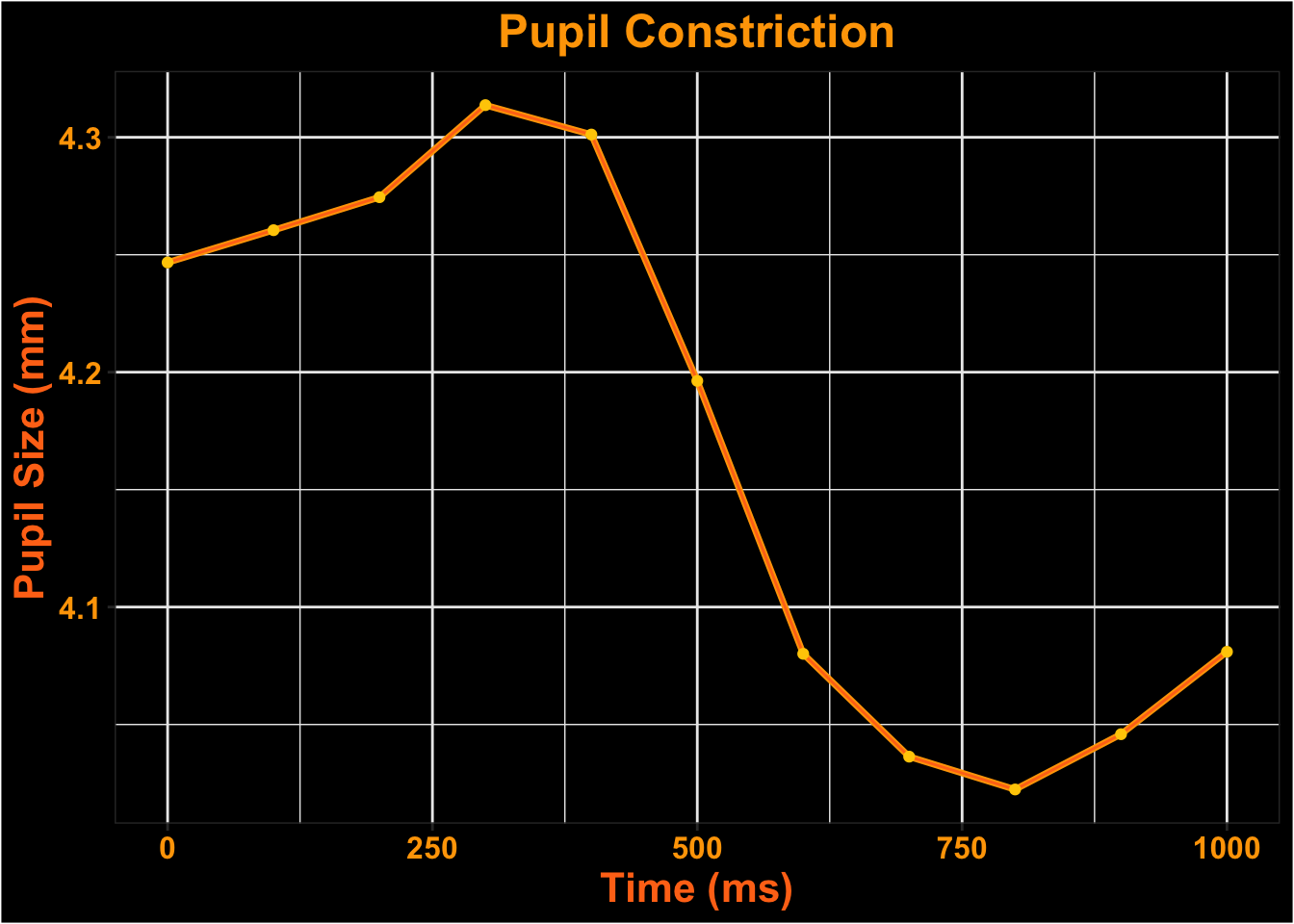
Visualize Time-course
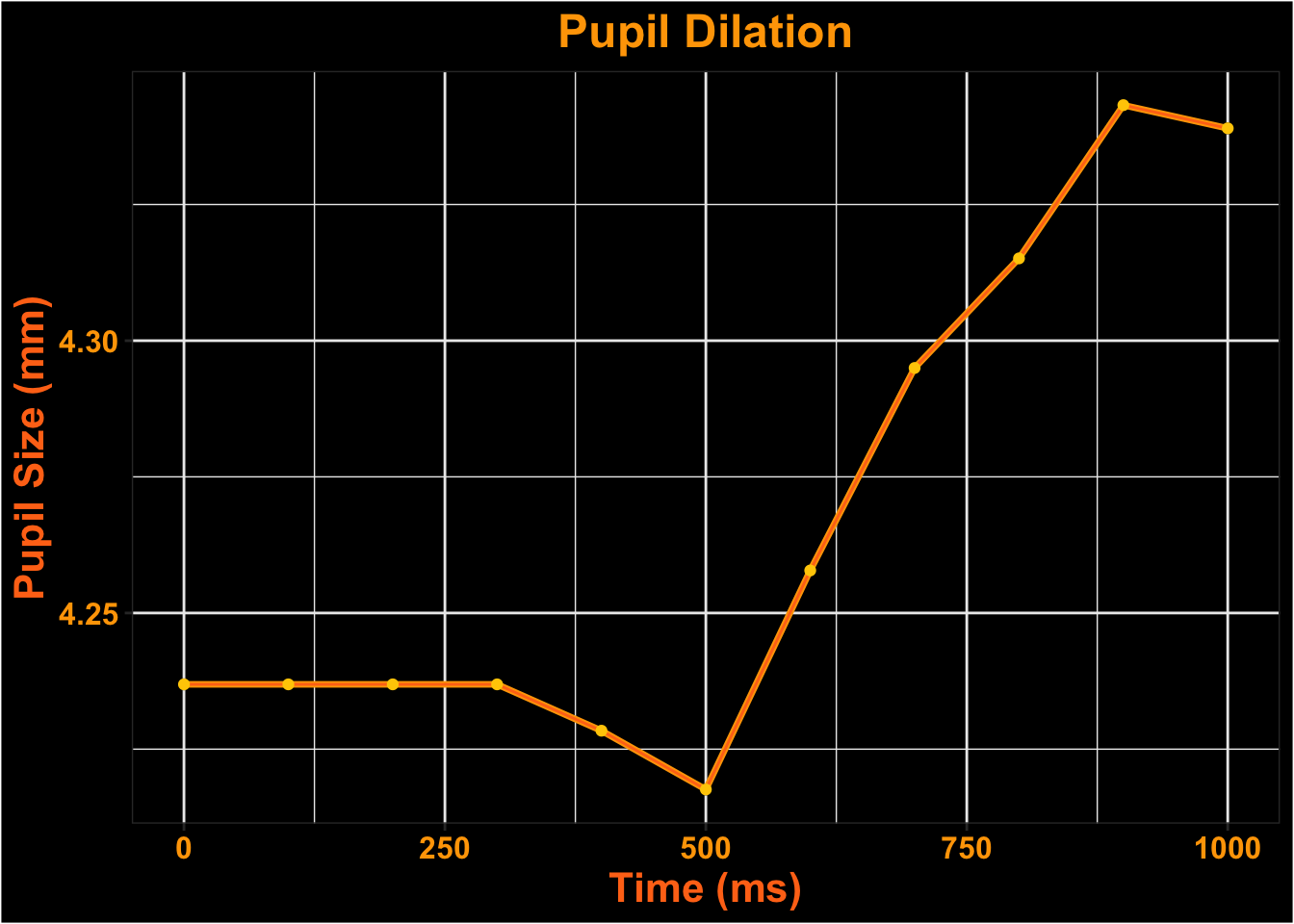
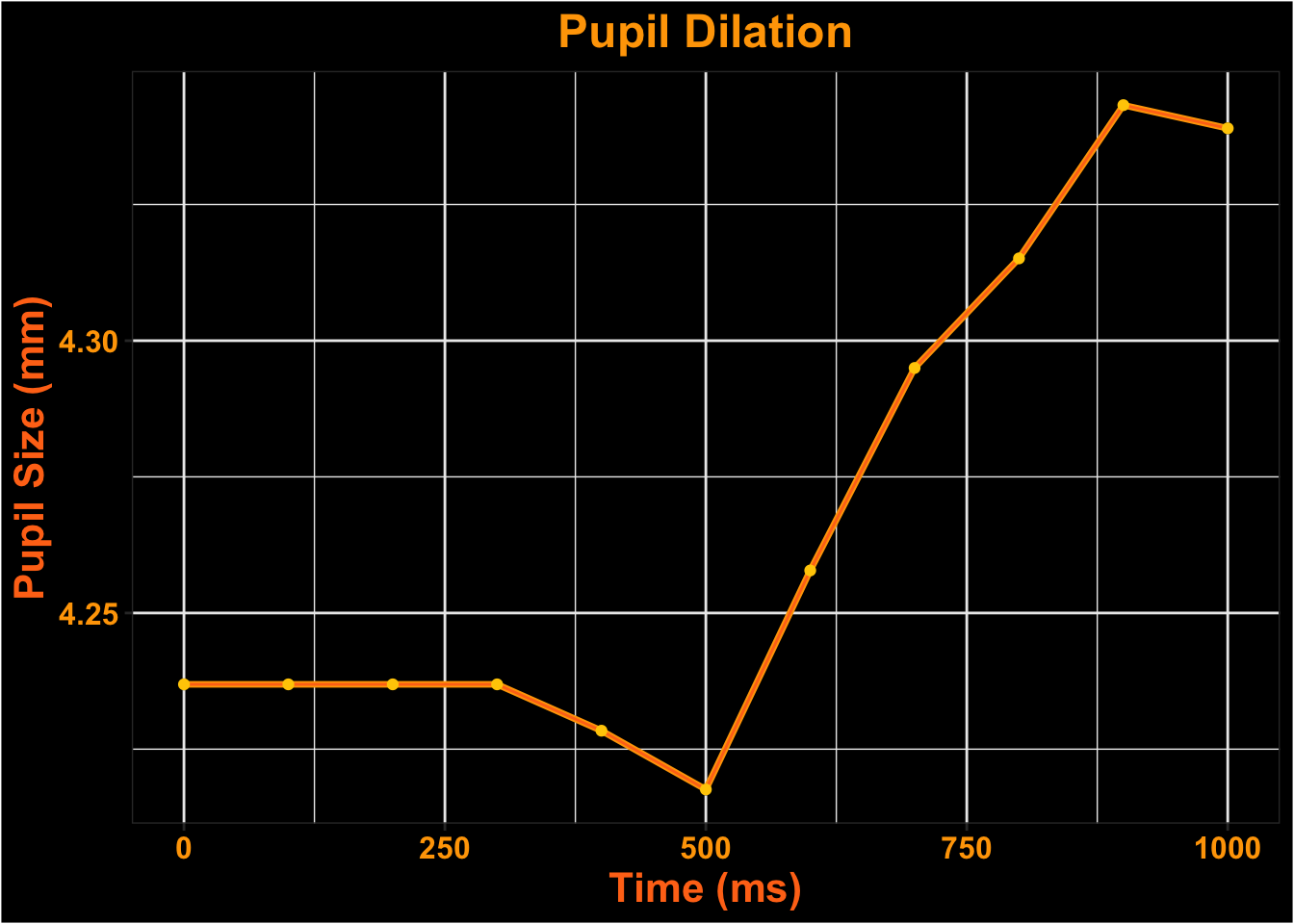
Surface Looks
What does the data look like if we constrain the analyses to only include looks to the laptop screen?
| subject | trial | time | gaze_detected_on_surface | gaze_position_on_surface_x_normalized | gaze_position_on_surface_y_normalized | pupil | blink | message | fixation_id |
|---|---|---|---|---|---|---|---|---|---|
| test1 | 1 | 0.000000 | TRUE | 0.503664 | 0.523080 | 4.28100 | FALSE | trial-started-light | 1 |
| test1 | 1 | 5.000000 | TRUE | 0.505029 | 0.524774 | 4.24610 | FALSE | 1 | |
| test1 | 1 | 9.999756 | TRUE | 0.505353 | 0.521651 | 4.22220 | FALSE | 1 | |
| test1 | 1 | 15.011963 | TRUE | 0.505129 | 0.522213 | 4.23325 | FALSE | 1 | |
| test1 | 1 | 20.000000 | TRUE | 0.503285 | 0.520440 | 4.27000 | FALSE | 1 | |
| test1 | 1 | 25.000000 | TRUE | 0.504079 | 0.509780 | 4.25900 | FALSE | 1 |
Let’s take a look at the data we have.
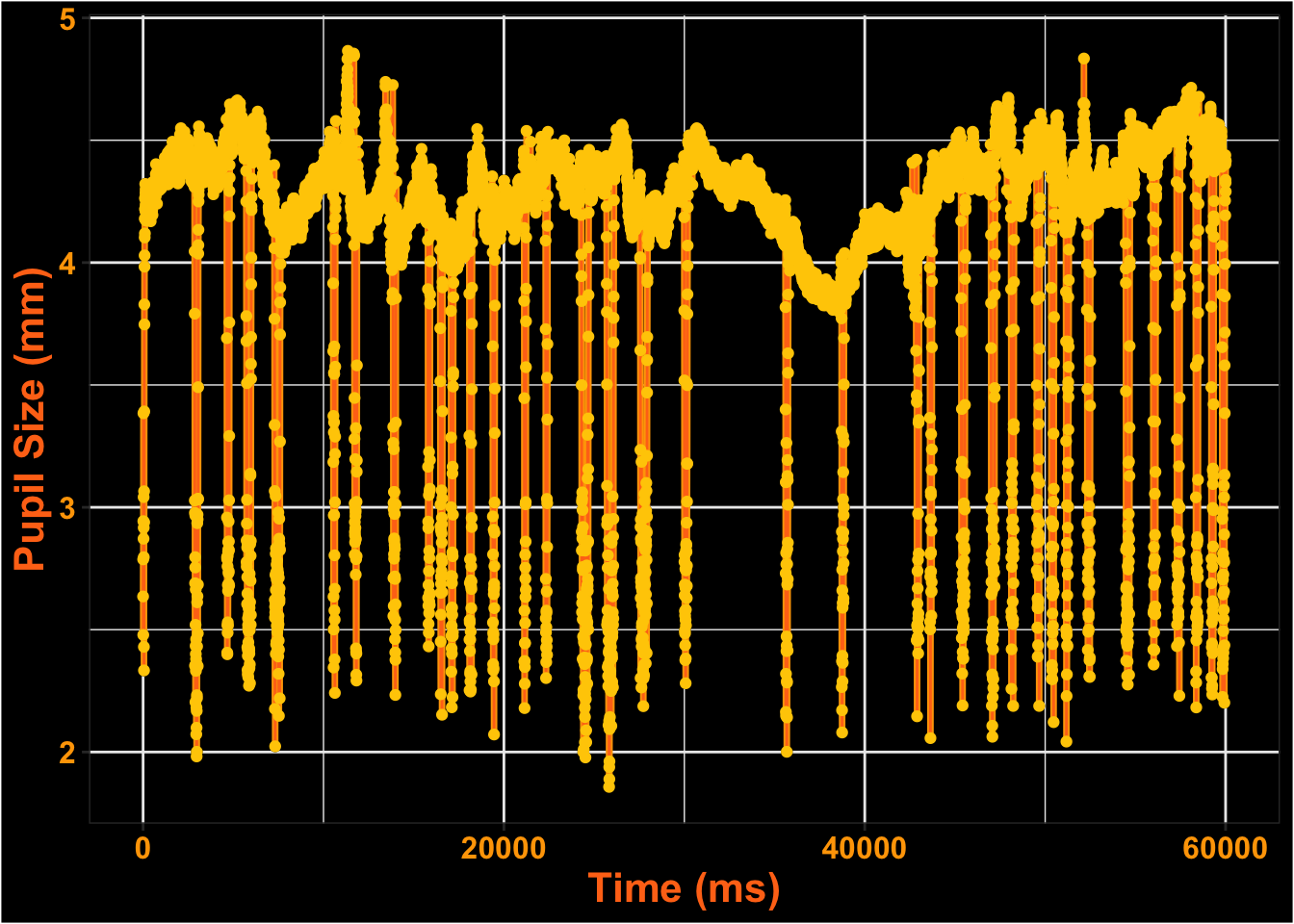
This is what the pupil data looks like for the entire time course.
Extending Blinks
We see dips in the pupil signal—these are most likely caused by blinks. Pupil Labs includes a blink detection algorithm, which we’ll use here. We first set pupil size values corresponding to blinks to NA, then use extend_blinks()to extend blinks 100 ms forward and backward in time.
Interpolate blinks
Next, we linearly interpolate over blinks and smooth the data using a 5-point moving average.
# Smooth and Interpolate
smooth_interp <- smooth_interpolate_pupil(pup_extend, pupil="pupil", extendpupil="extendpupil", extendblinks=TRUE, step.first="smooth", maxgap=Inf, type="linear", hz=200, n=5)Then, calculate missing data and remove trials with more than 50% missing data.
pup_missing<-count_missing_pupil(smooth_interp, missingthresh = .5)Unlikely Pupil Sizes
Now, keep only plausible pupil diameters between 2 mm and 9 mm.
pup_outliers<-pup_missing |>
dplyr::filter (pup_interp >= 2, pup_interp <= 9)MAD
Get rid of artifacts we might have missed during some earlier steps.
Onset
Let’s only look from the start of the trial until 1000 ms
Downsample
Downsample the time-course to 100 ms.
#downsample
timebins1<- downsample_gaze(pupil_onset, bin.length=100, pupil="pup_interp", timevar = "time", aggvars = c("subject", "trial", "timebins"), type="pupil")
kable(head(timebins1))| subject | trial | timebins | aggbaseline |
|---|---|---|---|
| test1 | 1 | 0 | 4.246710 |
| test1 | 1 | 100 | 4.260450 |
| test1 | 1 | 200 | 4.274502 |
| test1 | 1 | 300 | 4.313692 |
| test1 | 1 | 400 | 4.301169 |
| test1 | 1 | 500 | 4.196293 |
Visualize Time-course